Collecting Data From Different Resources
To do any meaningful research into the "Tasty Bacon Trait" our researcher must first collect a series of data from different resources. This can be a laborious task in itself. First, let's look at the sort of data our researcher might want to look at.
Individual Resources resources
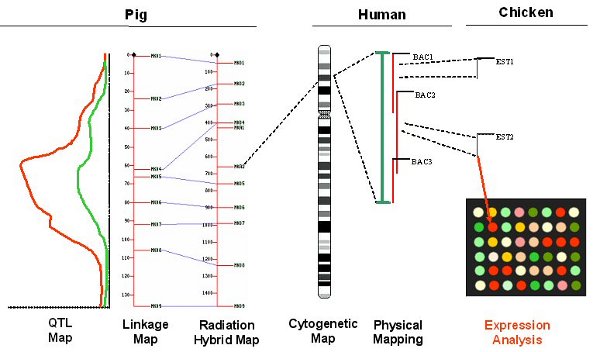
One resource contains a repository of genetic maps in pigs, which simply represent some identified traits in pigs as loci on a chromosome. This is a very qualitative study but gives our researcher a starting point, giving us this data.
Our researcher then finds another resource that contains other pig genetic maps, this time with some more statistical data that identifies some genes on the pig genome by location on a radiation hybrid map. Again, this doesn't give us any real statistical data but it gives us some candidate genes that might be associated with the trait.

Our researcher now knows that the region of the pig chromosome he's looking at is known to have homology with a region of one of the human chromosomes. He also knows that this region in human has been extensively sequenced, so he collects this data from a third database - his "Tasty Bacon" trait may have an ortholog in Homo Sapiens.

Now, our researcher thinks that some of these genes in human have homologs in chickens, and he knows of a resource that contains gene expression profiles for genes in chicken. He pulls some data down from this database so he can look for homologs between human and chicken genes, and get hold of the gene expression data for these homologs. In this way he can enrich his data with some more quantitative studies on gene function.
Finally, our researcher will need to access a fifth datasource and more to get hold of the publications associated with all of this data, and other studies that may be on the literature. There are many databases containing references to biological publications available so our researcher will need to spend some time querying these.
Correlating the Data correlation
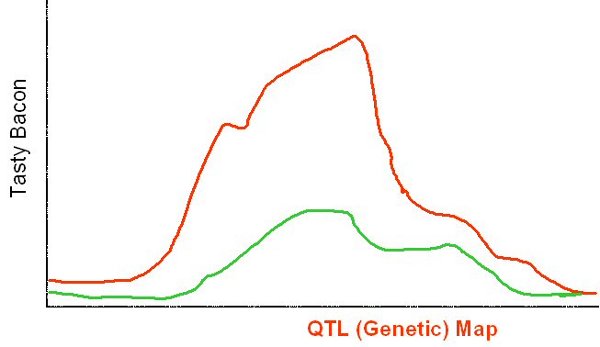
After our researcher has spent a considerable amount of time pooling data from these different resources, he then needs to correlate it by looking for overlaps, cross references, or sequence similarity. Maybe he runs some BLAST searches to work out which genes in his data are similar. This process may turn up new genes which he hadn't included before, so he might need to go back and fetch more data. After this trial-and-error process he may eventually come up with some statistically significant results, so he can draw a picture like this...
Adding value adding-value
After our researcher has obtained this overlay, he may be able to identify links by looking at genes near each other, so this consolidated view has added some new value. However, there is plenty of added value we'll miss out using these techniques.
Possibly, one resource contains some information about a sequence region that is incomplete, but another resource contains the "missing" data. Unfortunately, this would be missed using this strategy for fetching, but could possibly be retrieved if our databases lined up better - so they could "talk to each other". Also, the researcher probably has to search separately for the publications referring to this studies using native interfaces, so he doesn't have all his data ready to hand in his single view. These problems could be solved by representing the data, along with it's metadata, in a "Semantic Web" way. We''ll look at this next.

